Hello Readers,
In this post we examine at a specific hashtag on Twitter: #PublicHealth. When I am not spending time studying analytics, I concentrate on the biostatistics and public health, which introduced me to data science.
Analytics plays an important role in public health, as evidence based decisions rely on proper data gathering and analysis for their scientific rigor. Data and information is the power to understand, especially in our times of technology and communication. For example, Google tapped into their search results related to flu symptoms, and managed to model and predict flu trends, which I covered in these two posts. So using Twitter, we can see snippets of public health mentions in what people are tweeting.
Because Twitter is a real-time micro-blogging site, we can take a snapshot of tweets for a specific user, topic, or time-frame. So here I chose to exhibit the topic #PublicHealth, in light of the ebola epidemic spreading in Western Africa. This particular outbreak is one of worst in history, occurring in Sierra Leone, Guinea, and Liberia with 982 cases and 613 deaths (as of July 17) since March of 2014. The Centers for Disease Control and Prevention (CDC) has sent specialists to track, contain, and inform the locals about the Ebola outbreak.
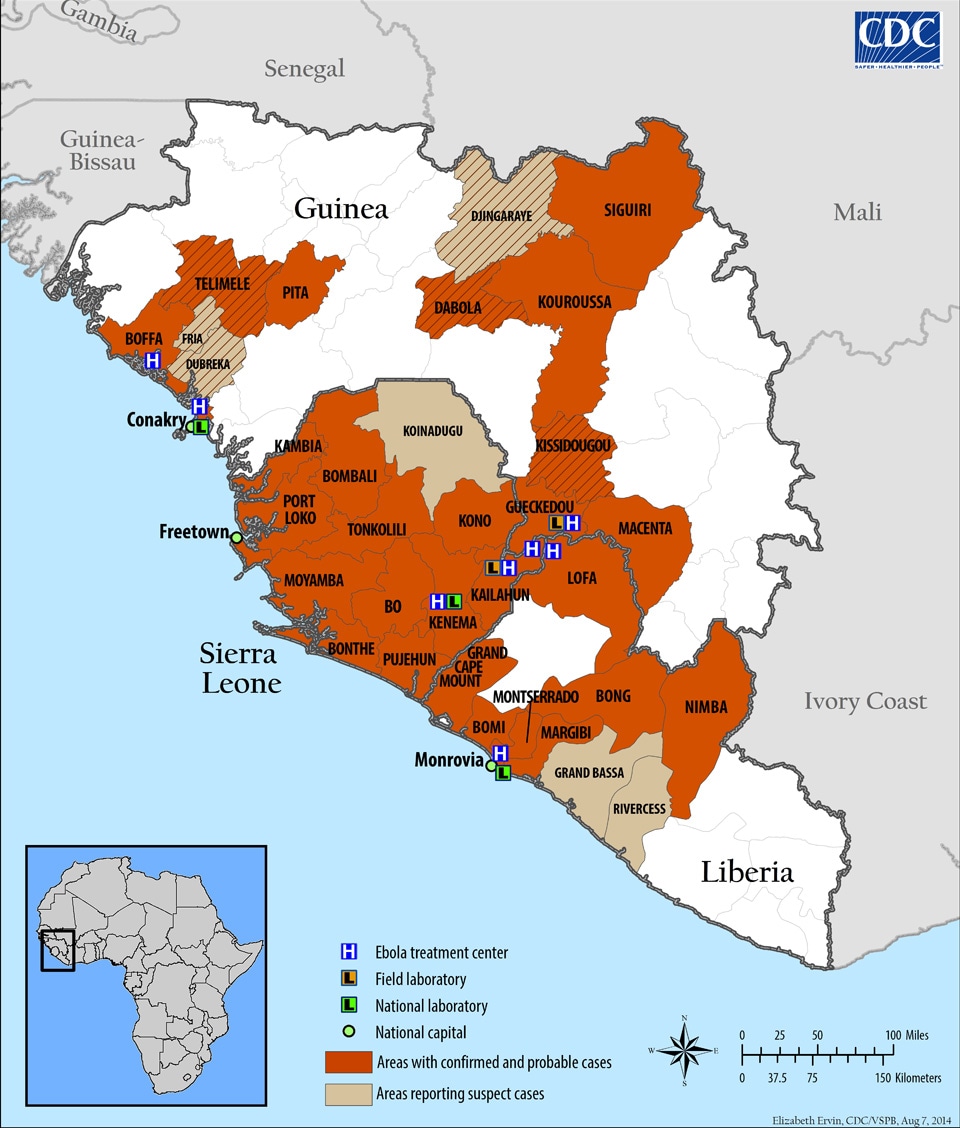 |
| Ebola Map, by CDC |
Returning back to Twitter and #PublicHealth, from the 300 tweets I queried in R, by using text transformation in R, I created a word cloud of the words in the tweets:
 |
| #Public Health |
The querying from Twitter, text transformation, and word cloud creation code can be found in the Appendix, below. As we investigate the word cloud, aside from "publichealth", we see some terms, such as:
- "croakeyblog", a blog about health,
- "profkevinfenton", Dr. Kevin Fenton, director of health and wellbeing at Public Health England,
- "data",
- "human",
- "ecigarettes",
- "healthcare",
- and "ebola" on the top left.
#PublicHealth covers a wide range of subtopics, and also links to many others as well. With increased globalization, and aging populations, both communicable and non-communicable diseases are rising across the world. Here is the World Health Organization (WHO) on immunizations, and reducing preventable deaths.
For those interested in studying or learning more about public health, consider the Johns Hopkins Bloomberg School of Public Health, and the Johns Hopkins Public Health Studies
undergraduate program.
Thanks for reading,
Wayne
@beyondvalence
Code Appendix:
Text Transformation Code:1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 | > # load library > library(tm) > # > # transforming function (by Wayne) > # where x is text and w.stop is stopword vector, > # w.keep are words to remove from stopwords > # Term-Doc default, if false, then Doc-Term > transform.text <- function(x, w.keep=c(""), w.stop=c(""), TD=TRUE) { + + cat("Data has ", length(x), " documents.\n") + + cat("Beginning text transformation: \n\n") + + cat("Transforming to Corpus... (1/6)\n") + text <- x + text.corpus <- Corpus(VectorSource(text)) + + cat("Formatting Corpus... (2/6)\n") + # lower case + text.corpus <- tm_map(text.corpus, tolower) + # remove punctuation + text.corpus <- tm_map(text.corpus, removePunctuation) + # remove numbers + text.corpus <- tm_map(text.corpus, removeNumbers) + # remove URLs + removeURLs <- function(x) gsub("http[[:alnum:]]*", "", x) + text.corpus <- tm_map(text.corpus, removeURLs) + # add stopwords w + myStopWords <- c(stopwords('english'), w.stop) + # remove vector w from stopwords + myStopWords <- setdiff(myStopWords, w.keep) + # remove stopwords from corpus + text.corpus <- tm_map(text.corpus, removeWords, myStopWords) + + cat("Stemming Words... (3/6)\n") + # keep corpus copy for use as dictionary + text.corpus.copy <- text.corpus + # stem words #### + text.corpus <- tm_map(text.corpus, stemDocument) + + cat("Completing Stems... (4/6)\n") + # stem completion #### + text.corpus <- tm_map(text.corpus, stemCompletion, + dictionary=text.corpus.copy) + + if(TD==TRUE) { + cat("Creating T-D Matrix... (5/6)\n") + text.TDM <- TermDocumentMatrix(text.corpus, + control=list(wordLengths=c(1,Inf))) + cat("Text Transformed! (6/6)\n\n") + return(text.TDM) + } else { + cat("Creating D-T Matrix... (5/6)\n") + # create Doc-Term #### + text.DTM <- DocumentTermMatrix(text.corpus, + control=list(wordLengths=c(1, Inf))) + cat("Text Transformed! (6/6)\n\n") + return(text.DTM) + } + + } |
Retrieving #PublicHealth Tweets Code:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 | > library(twitteR) > library(tm) > > # load twitter cred #### > load("cred.Rdata") > registerTwitterOAuth(cred) [1] TRUE > > # configure RCurl options > RCurlOptions <- list(capath=system.file("CurlSSL", "cacert.pem", package = "RCurl"), + ssl.verifypeer = FALSE) > options(RCurlOptions = RCurlOptions) > > # query twitter for #PublicHealth in tweets, n=300 #### > pH <- searchTwitter("#PublicHealth", n=300, lang="en", + cainfo=system.file("cacert.pem")) > save(pH, file="publicHealth.rdata") > > # to data.frame #### > pH.df <- do.call("rbind", lapply(pH, as.data.frame)) > # use textTransformation function #### > pH.tdm <- transform.text(pH.df$text, w.stop = c("amp", "rt") ,TD = TRUE) Data has 300 documents. Beginning text transformation: Transforming to Corpus... (1/6) Formatting Corpus... (2/6) Stemming Words... (3/6) Completing Stems... (4/6) Creating T-D Matrix... (5/6) Text Transformed! (6/6) > # find terms with n > 20 [1] "advice" "blogs" "can" "climate" [5] "cost" "croakeyblog" "data" "day" [9] "elderly" "eye" "falls" "health" [13] "heat" "heatwave" "helping" "herts" [17] "india" "issue" "jimmcmanusph" "keep" [21] "major" "need" "neighbours" "pheuk" [25] "please" "prevent" "profkevinfenton" "publichealth" [29] "stories" "support" "today" "vulnerable" > |
Creating the Word Cloud Code:
1 2 3 4 5 6 7 8 9 10 11 12 13 | > # generate word cloud #### > library(wordcloud) Loading required package: Rcpp Loading required package: RColorBrewer > pH.matrix <- as.matrix(pH.tdm) > wordFreq.sort <- sort(rowSums(pH.matrix), decreasing=T) > # wcloud > set.seed(1234) > grayLevels <- gray( (wordFreq.sort + 10) / (max(wordFreq.sort) + 10)) > wordcloud(words=names(wordFreq.sort), freq=wordFreq.sort, + min.freq=3, random.order=F, colors=grayLevels) > |
No comments:
Post a Comment