Hello Readers,

Hope you guys are having a wonderful holiday! (I am.) Today in this post we will cover the k-means clustering technique in R.
We will use the familiar iris data set available in R.
Let us get started!
K-Means Clustering
Start by loading the cluster package by library(cluster) in R. The first and very important step in k-means clustering occurs when choosing the number of final clusters (k).
Therefore the k-means clustering process begins with an educated 'guess' of the number of clusters. With the k number of clusters, R selects k observations in the data to serve as cluster centers. Then the Euclidean distance is calculated from observations to the cluster centers, and the observations are placed in the cluster to which they are closest.
Then the center of each cluster is recalculated and the Euclidean distance is taken for each observation and the new cluster center. R checks every observation to see if it is closer to another cluster center and reassigns it if it is closer to another cluster. The process of center cluster recalculation and observation distance checking is repeated until observations stay in the same cluster.
The kmeans() function requires the choosing k observations as the centers of the clusters.
Before clustering, remove the species column from the iris data set to retain the numerical values only.
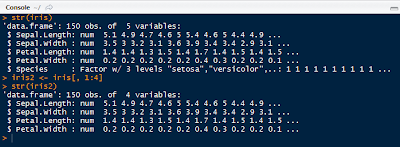 |
| Iris2 Data Set Without Species |
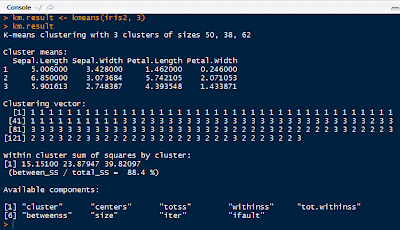 |
| K-Means Result |
kmeans() seeks to minimize the withinss for each cluster, which is the sum of of squared error (SSE) or scatter. It is calculated by taking the sum of the squares of the distances between the observations and centroid of each cluster.
To see how well the k-means clustering performed, we can create a table with the actual species of iris and the cluster numbers:
 |
| Species and Cluster Comparison |
As we can see, the clustering successfully clustered all the setosa species, but had difficulty with virginica (14 off) and versicolor (2 off). To quantify the agreement we can use the the library(flexclust) package, as shown below:
 |
| Adjusted Rank Index |
An agreement of 0.73 is not too bad, as it ranges from -1 (no agreement) to 1 (perfect agreement). We have a 0.73 agreement between the iris species and cluster solution.
Plotting Clusters
Next we can visualize the clusters and their centers we constructed, with the code below:
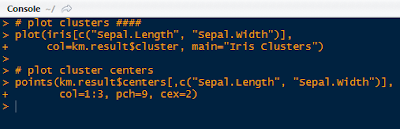 |
| Plotting Code |
And that yields the visual below. Note that the three diamonds are the cluster centers in black, green and red.
 |
| Iris Clusters Plot |
In the next Cluster Analysis post I will discuss finding a suitable k to begin the k-means analysis. In this case, we knew we had 3 species to begin, so it was easy to plug in the k. However, we will look at the within sum of squares and sets of criteria to see what k we will use for a data set on wine. Stay tuned!
Thanks for reading,
Wayne
@beyondvalence
No comments:
Post a Comment